Initial Results
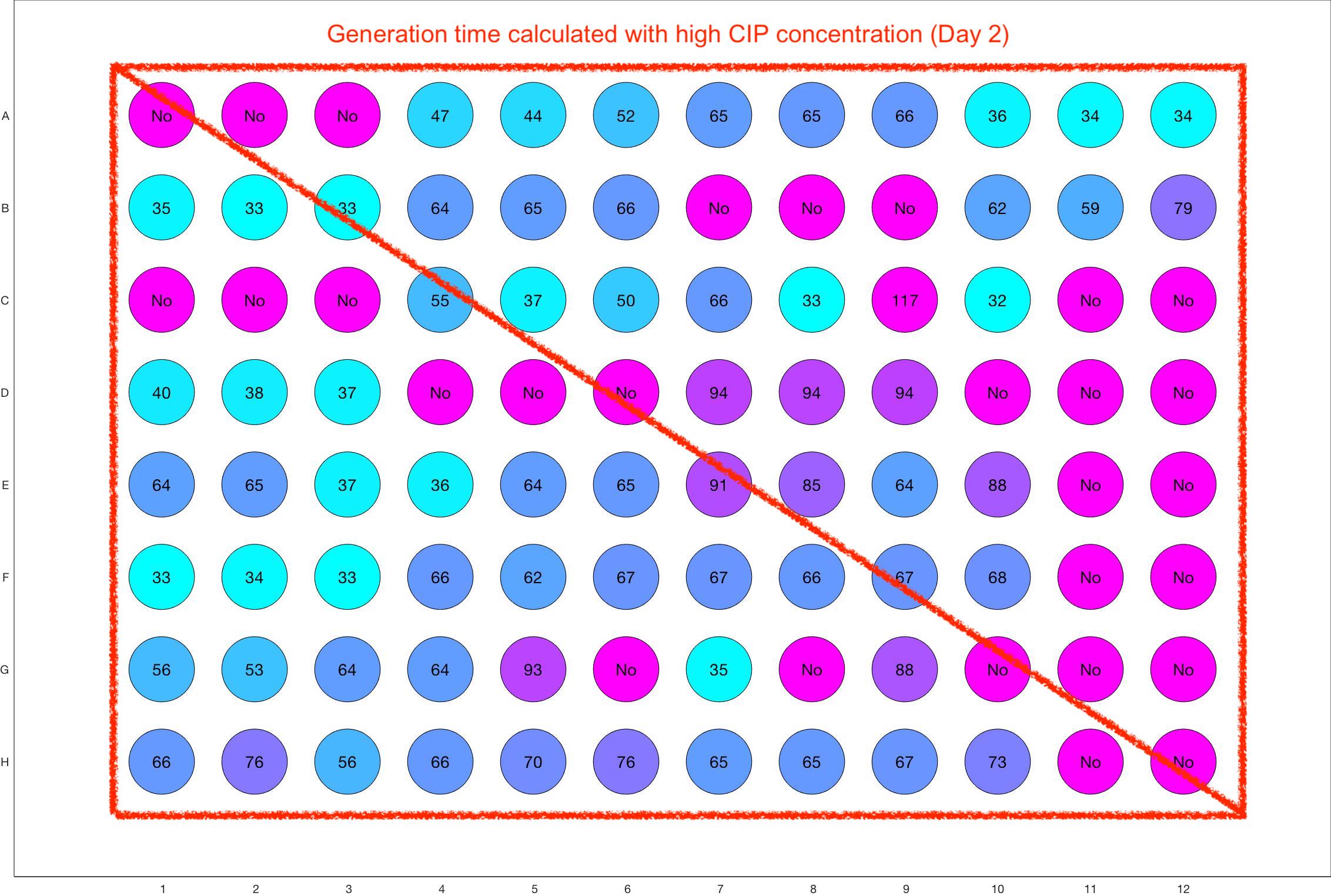
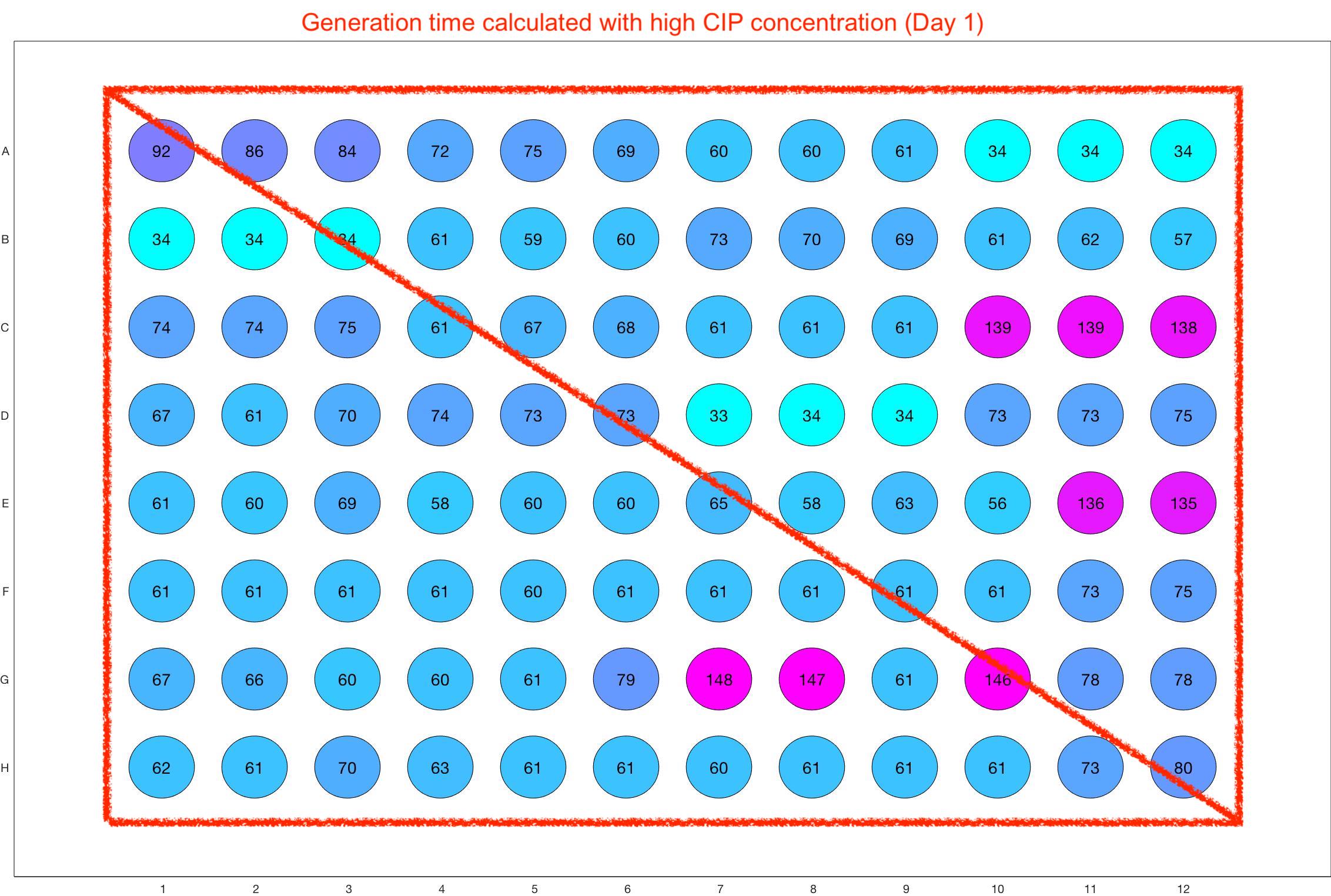
Monitoring bacterial growth on different antibiotics concentrations allows us to infer what is the minimum inhibitory concentration (MIC) for each antibiotic type. Antibiotic concentration just below the MIC are a good starting point for the experimental evolution. In these examples, we defined the MIC as the concentration with a doubling time above 100 minutes. The initial concentrations chosen for the evolution experiment are marked in RED.
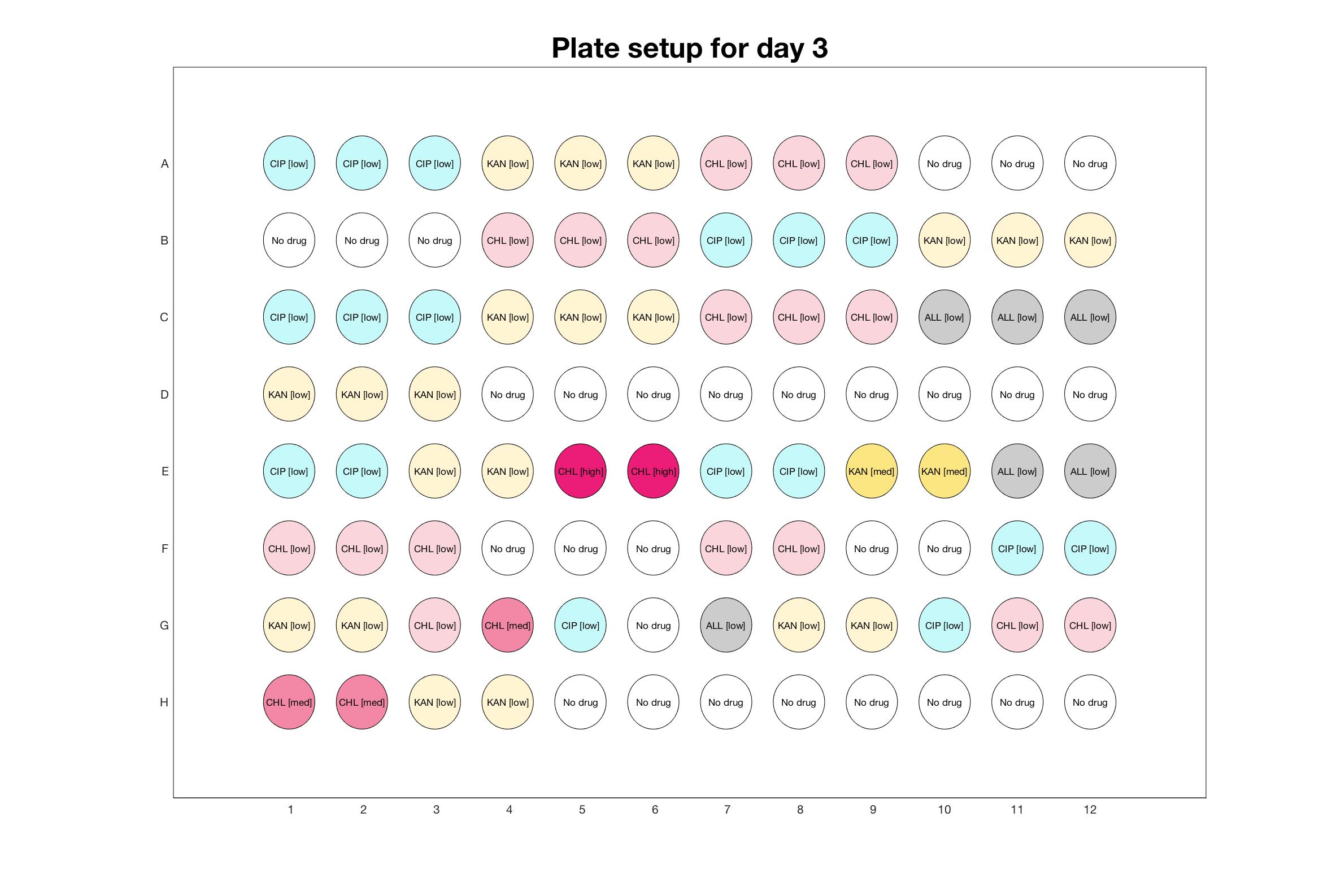
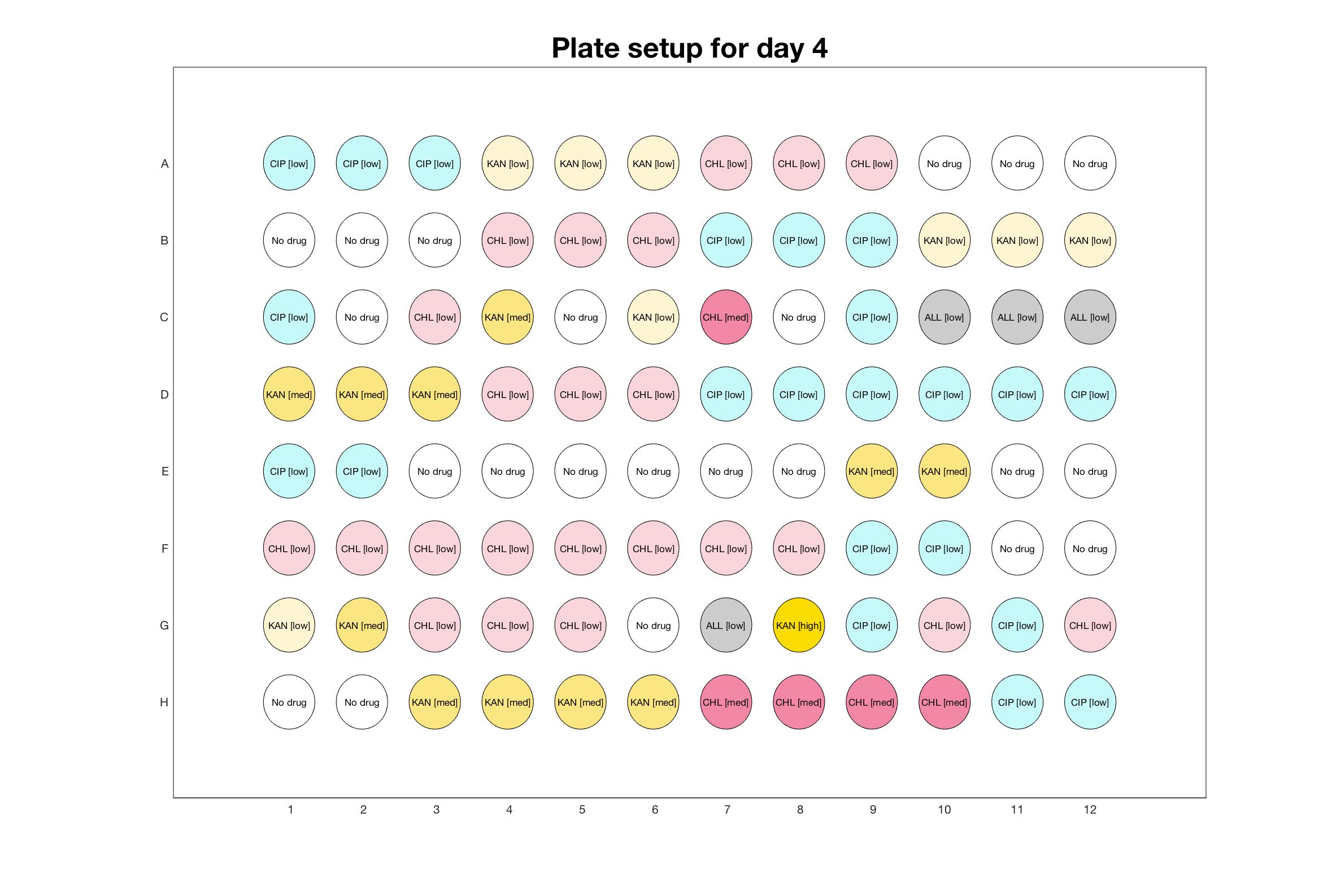
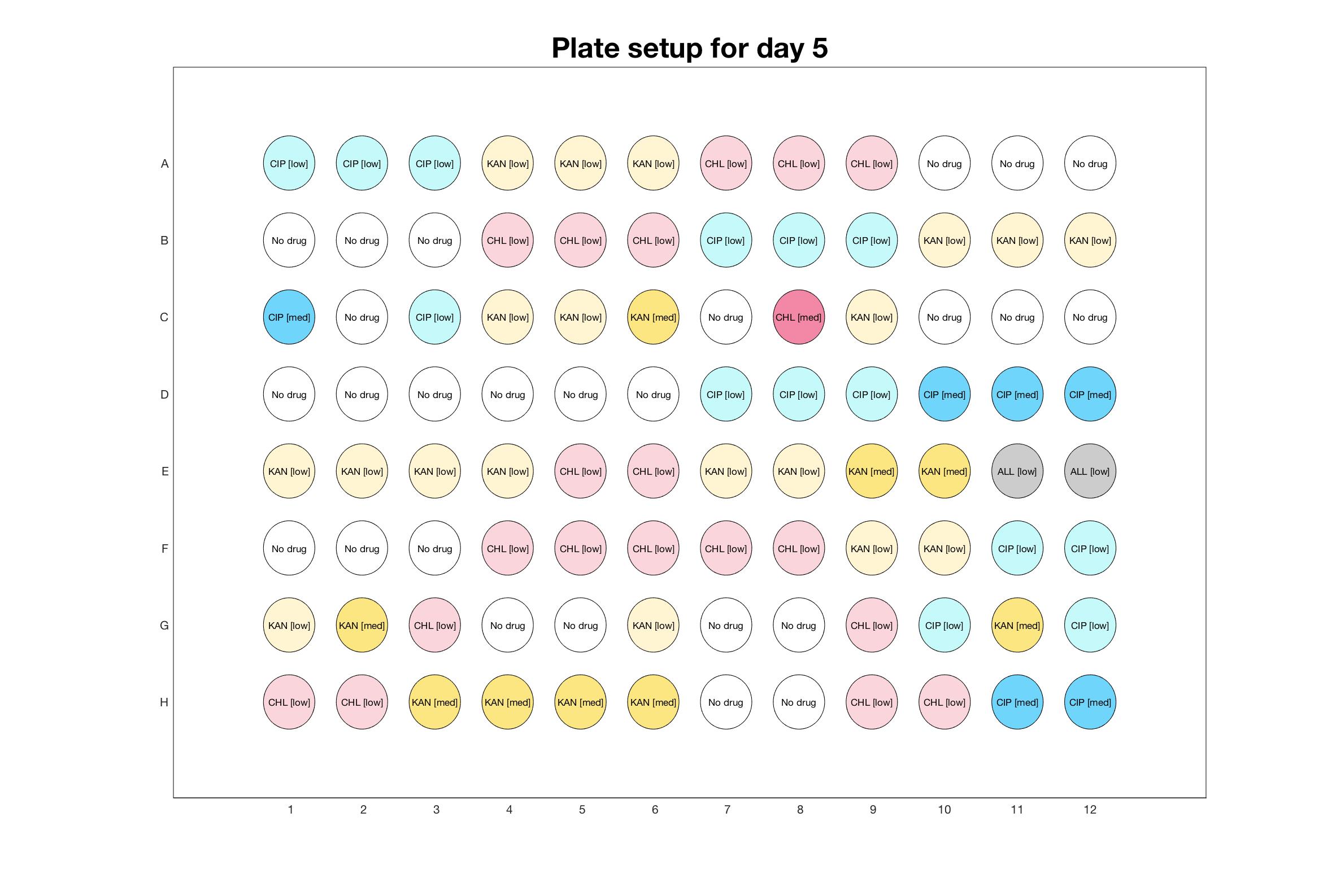
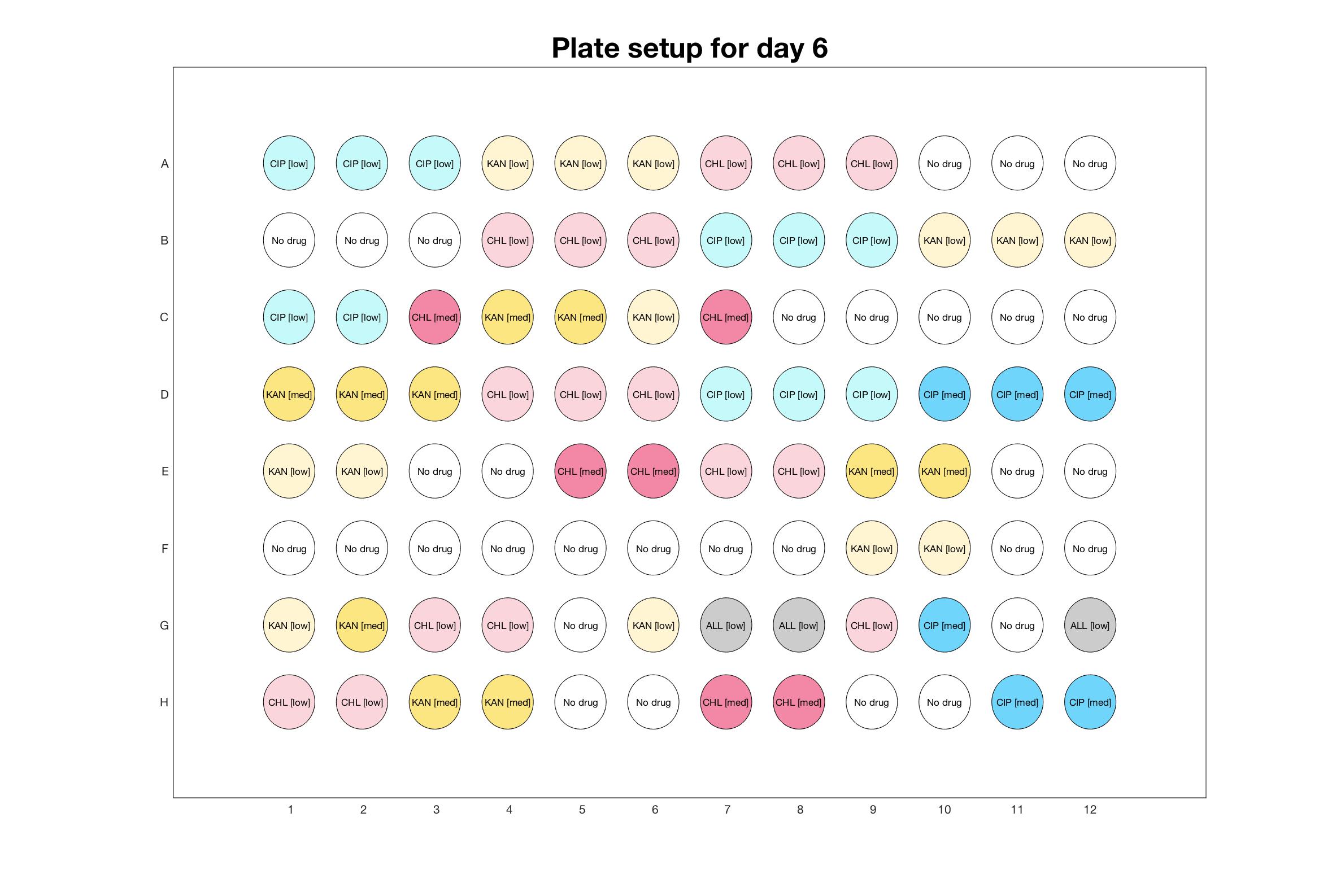
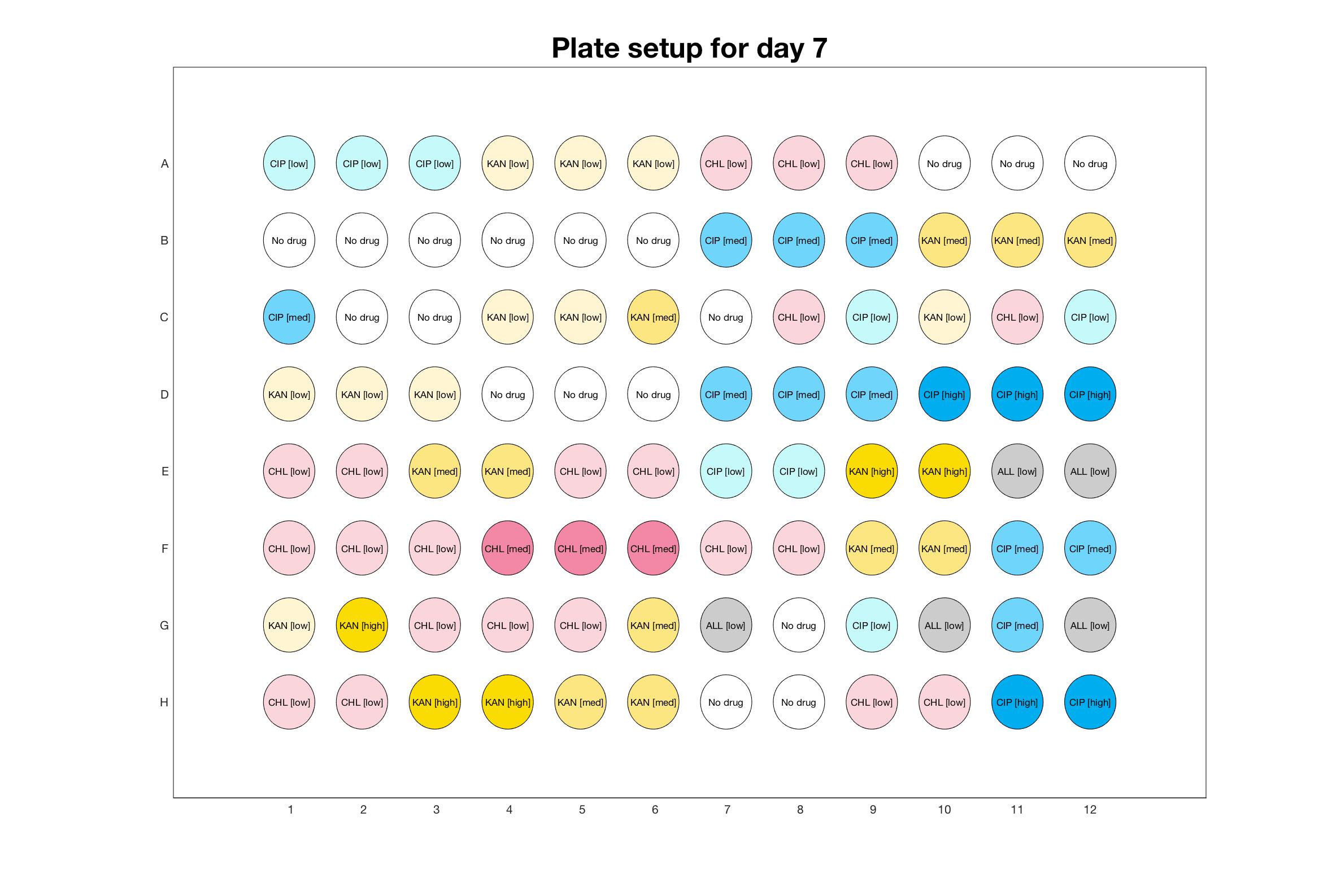
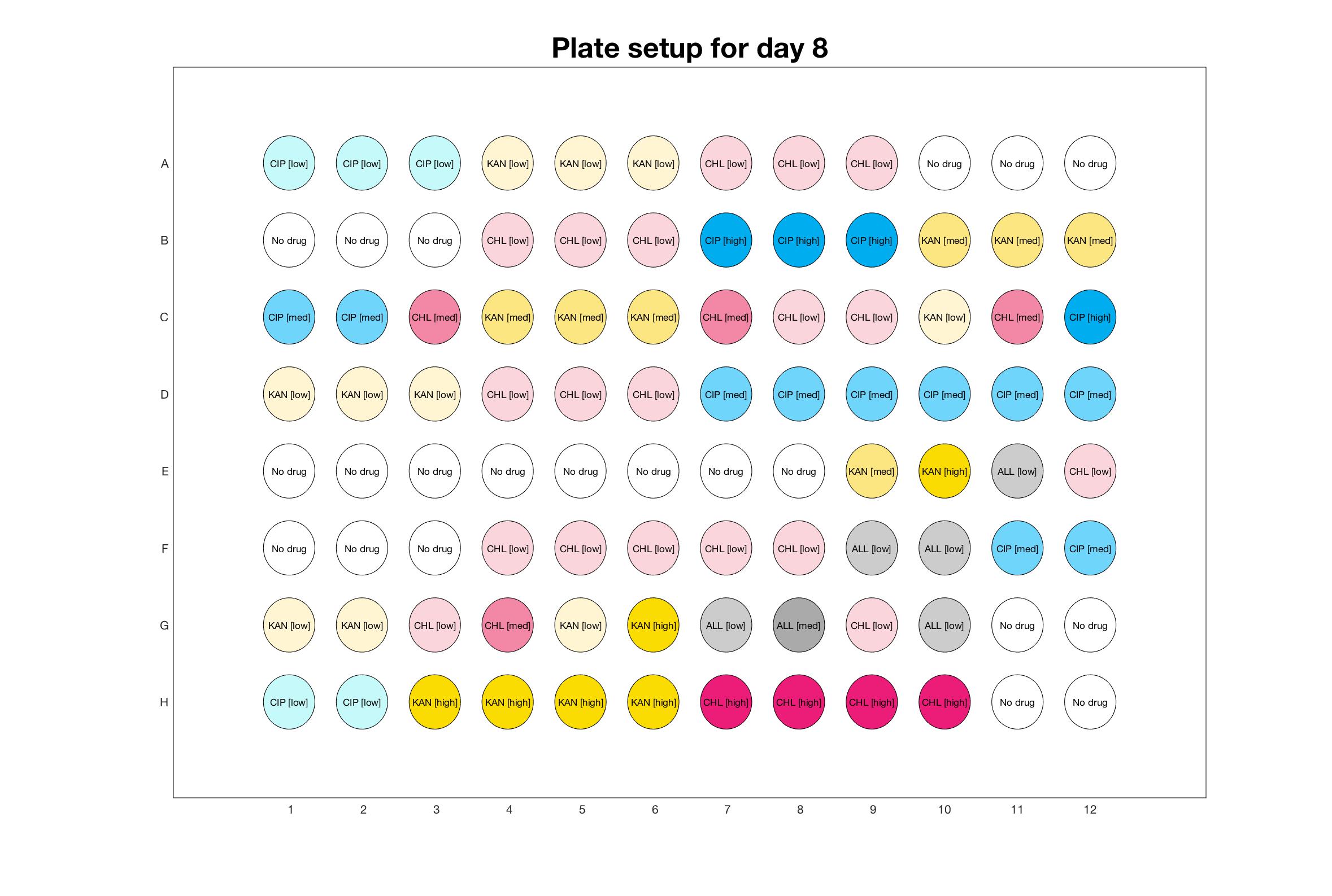
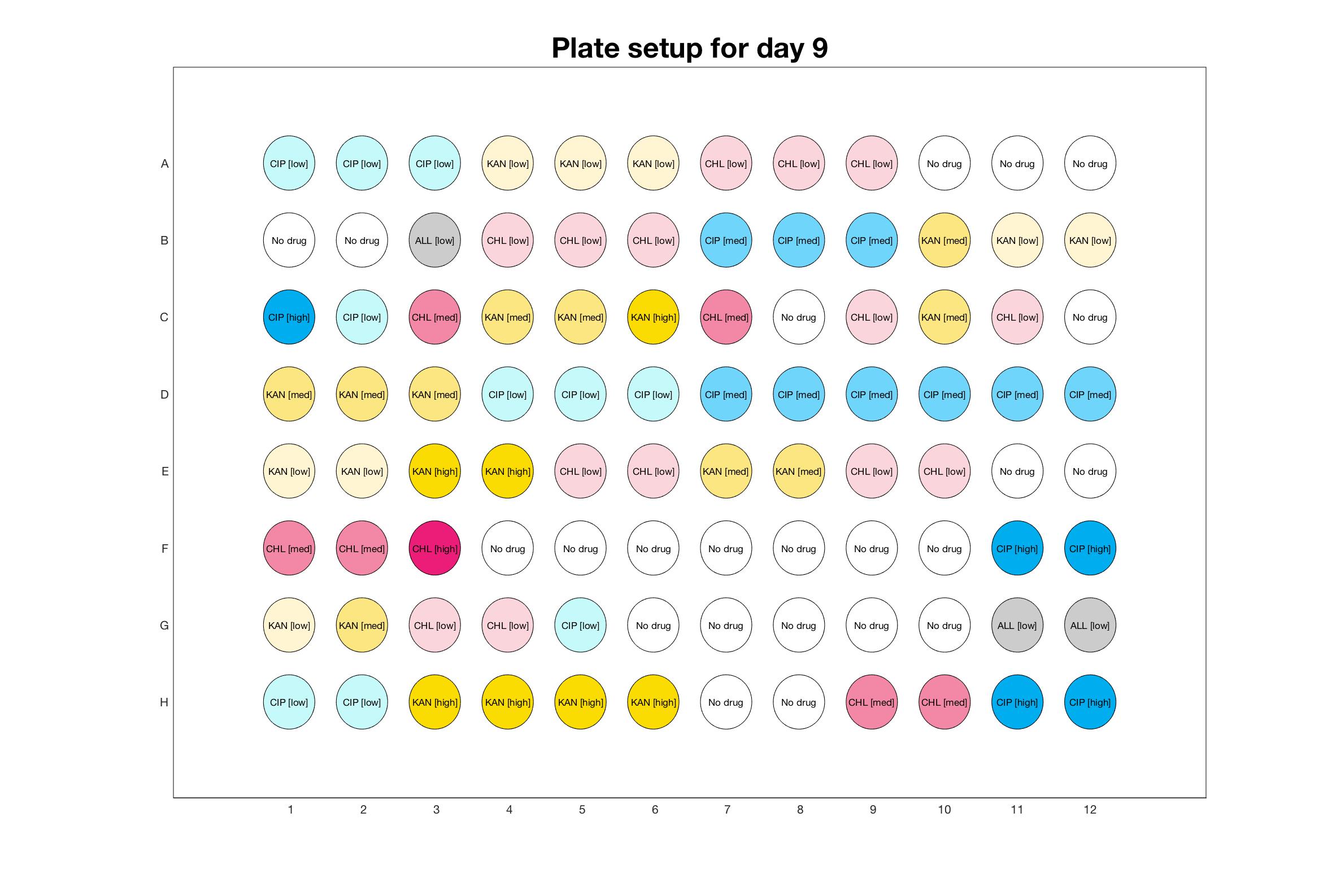
Daily drug regimen



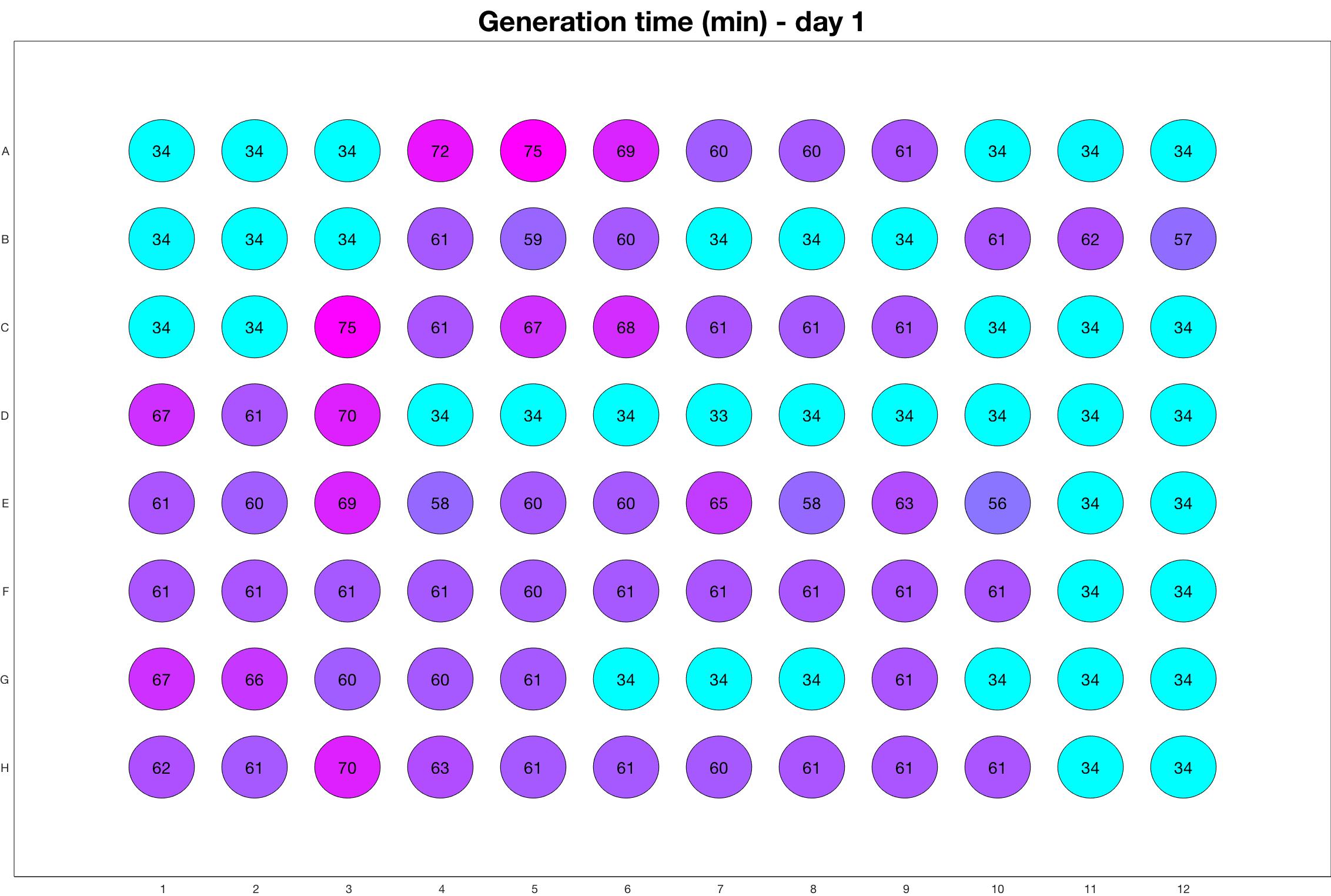
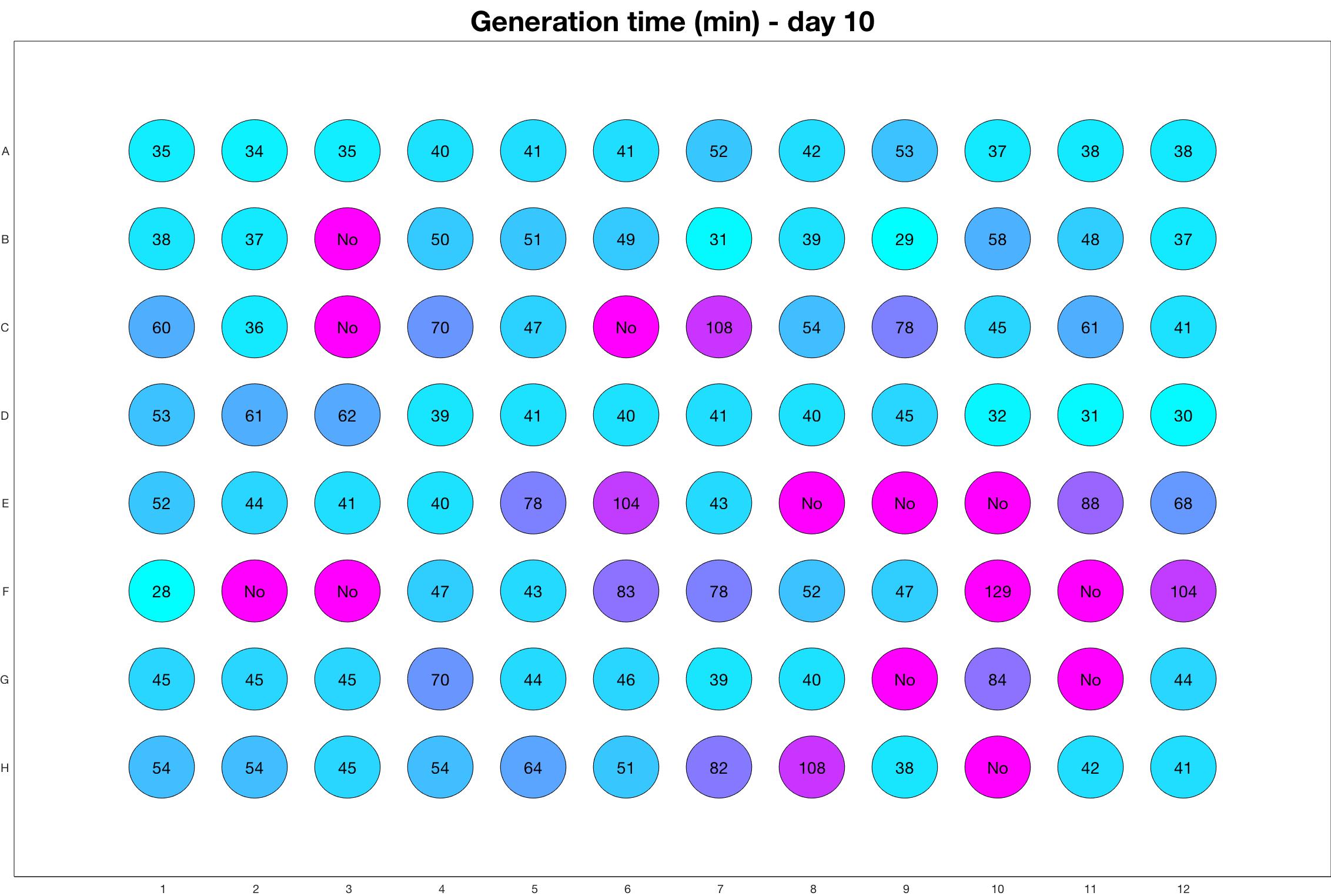
Daily observed generation time
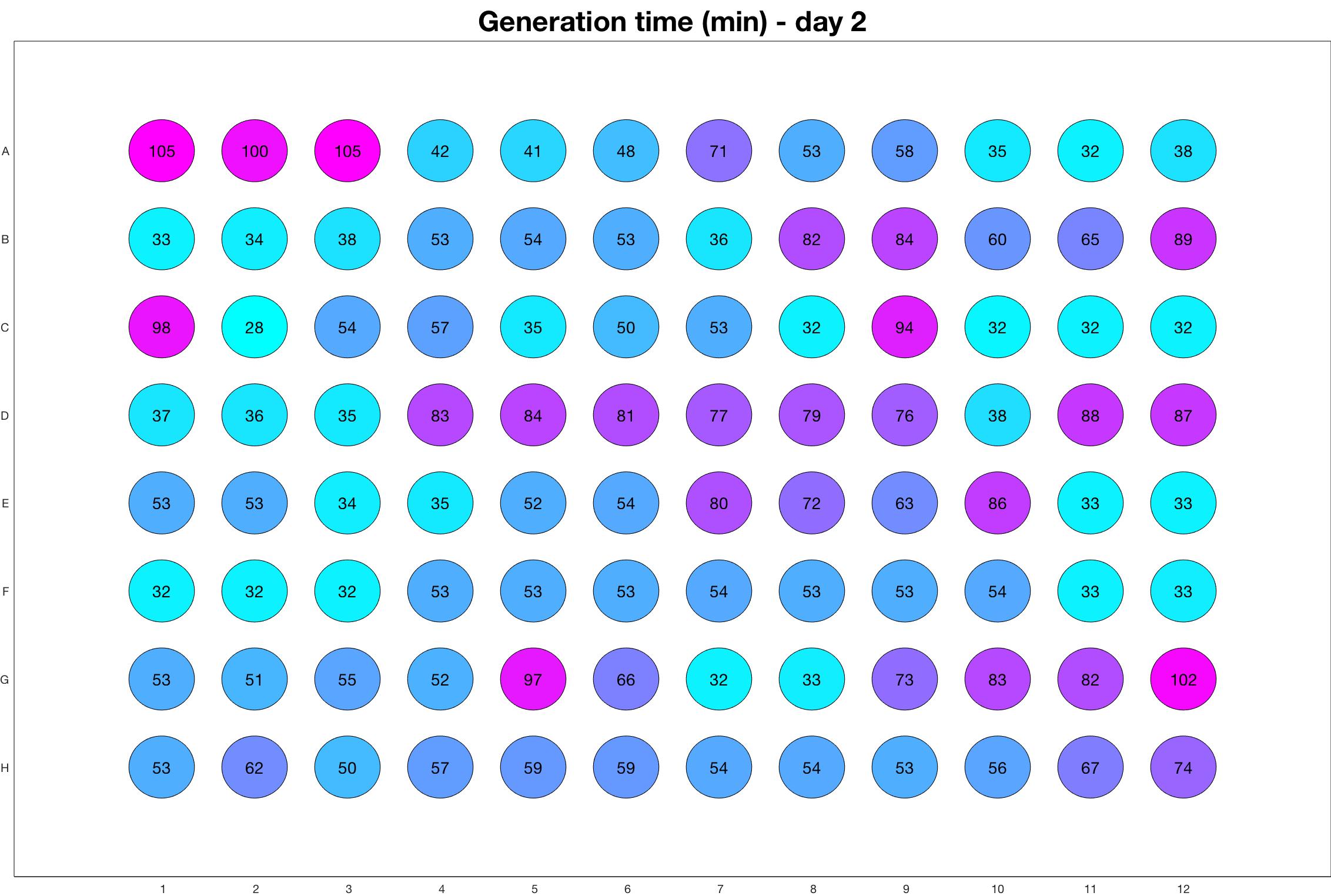
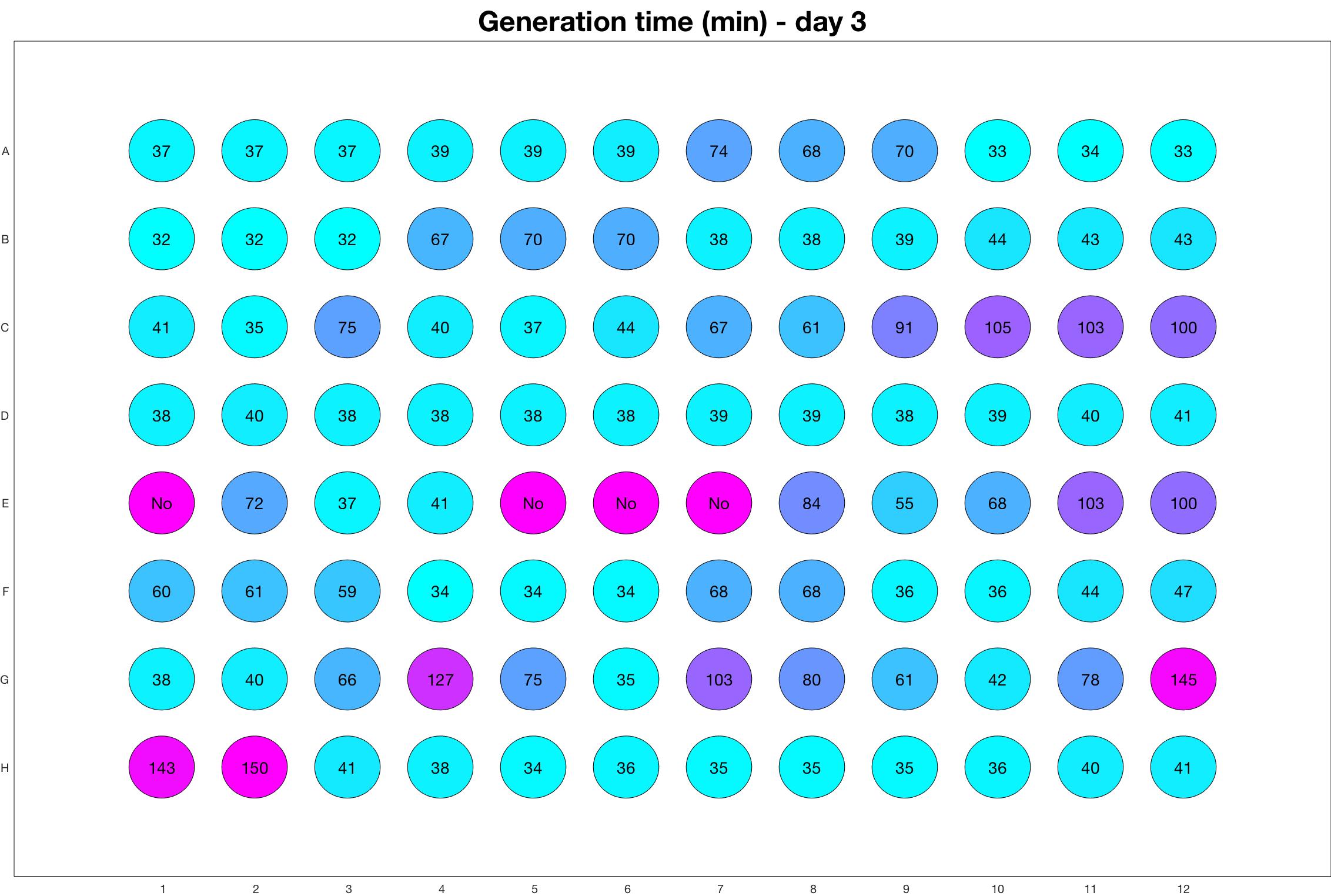
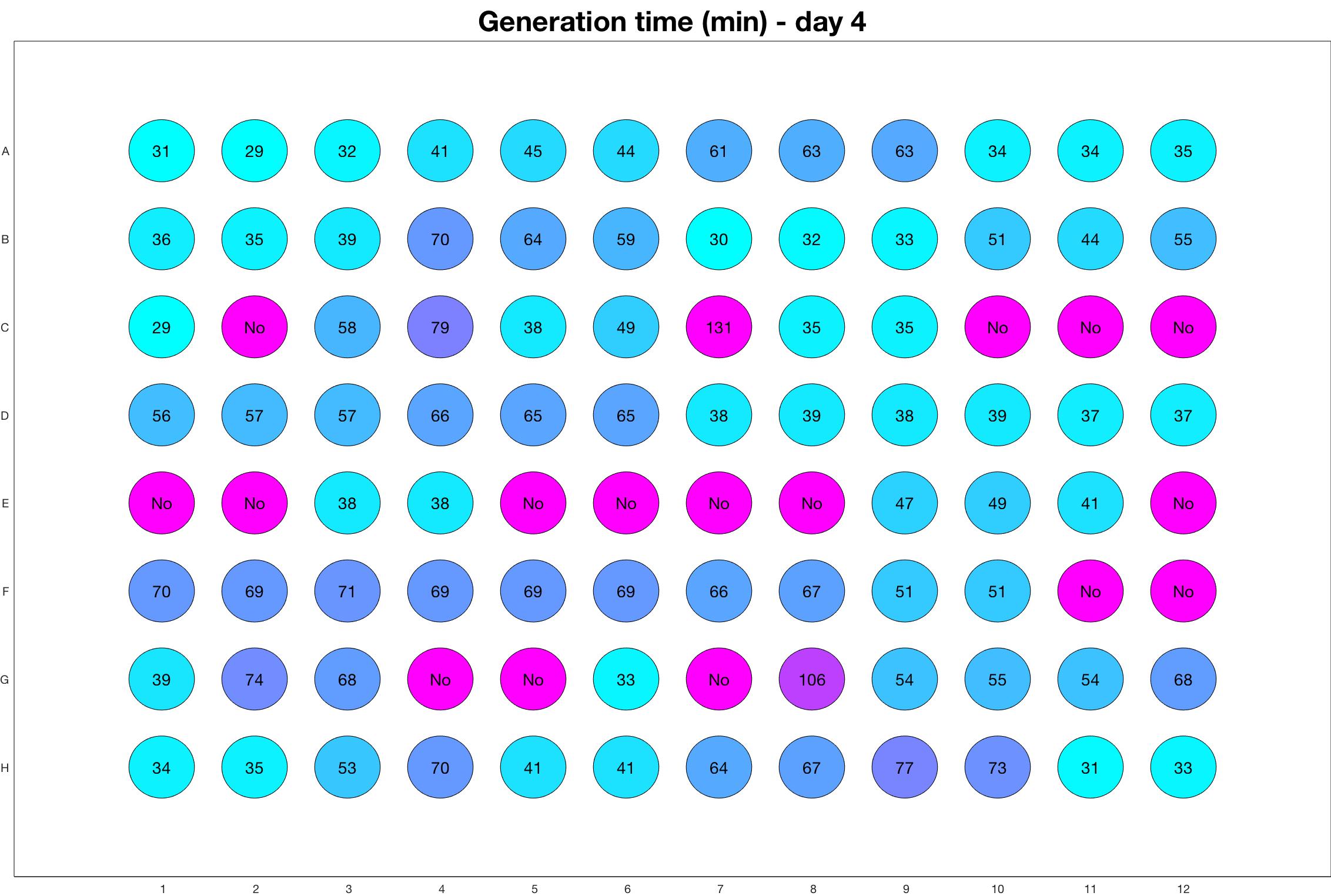
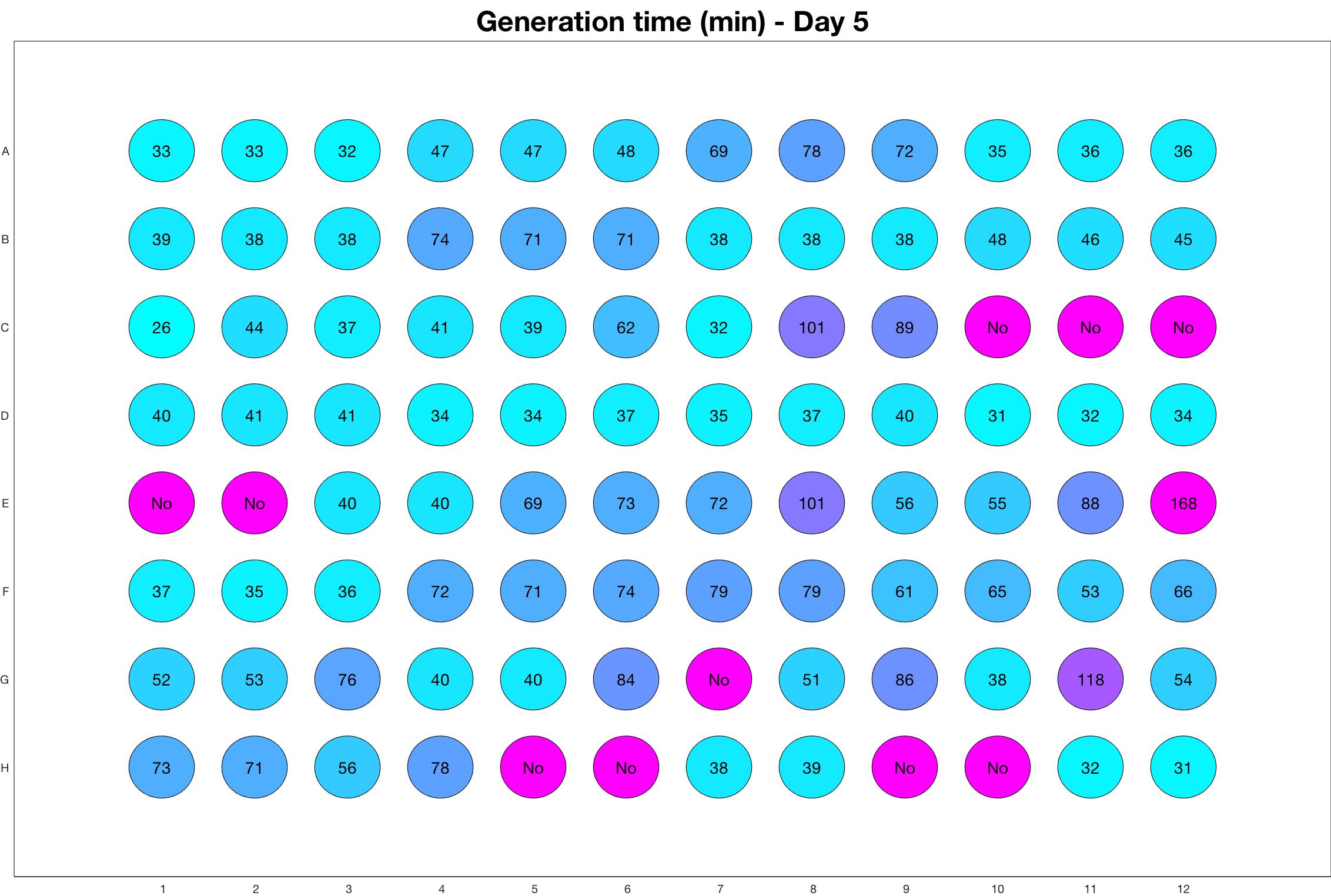
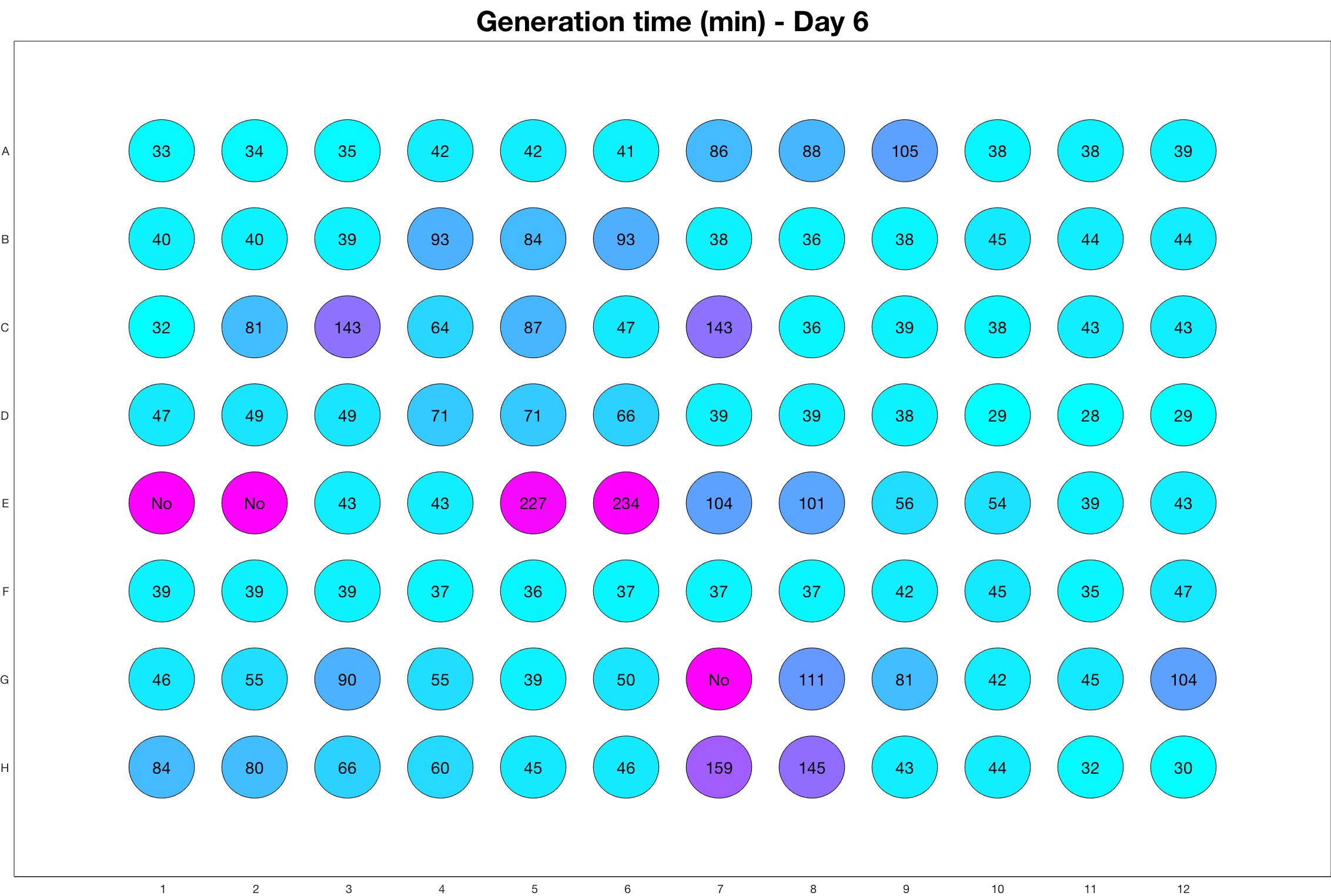
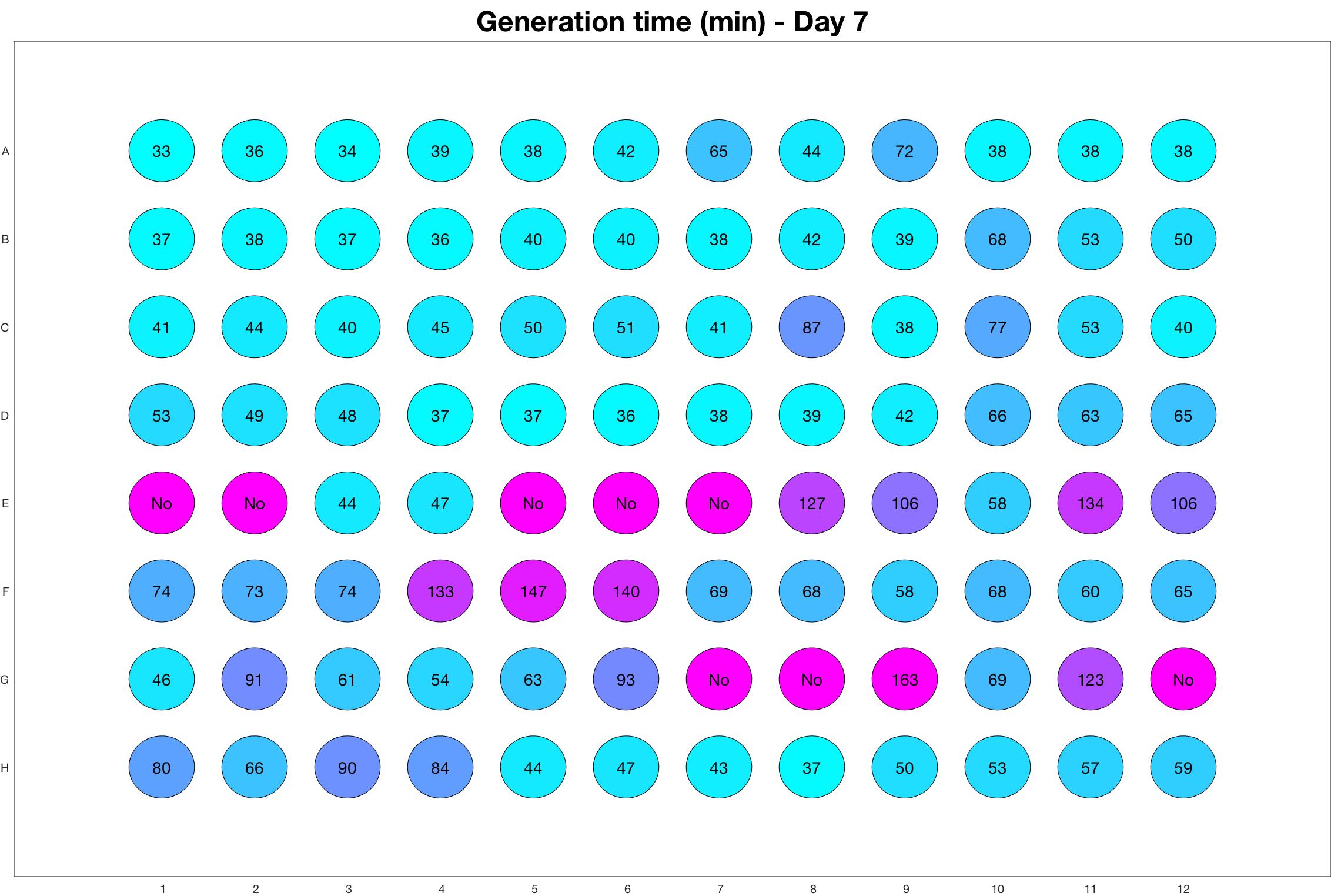
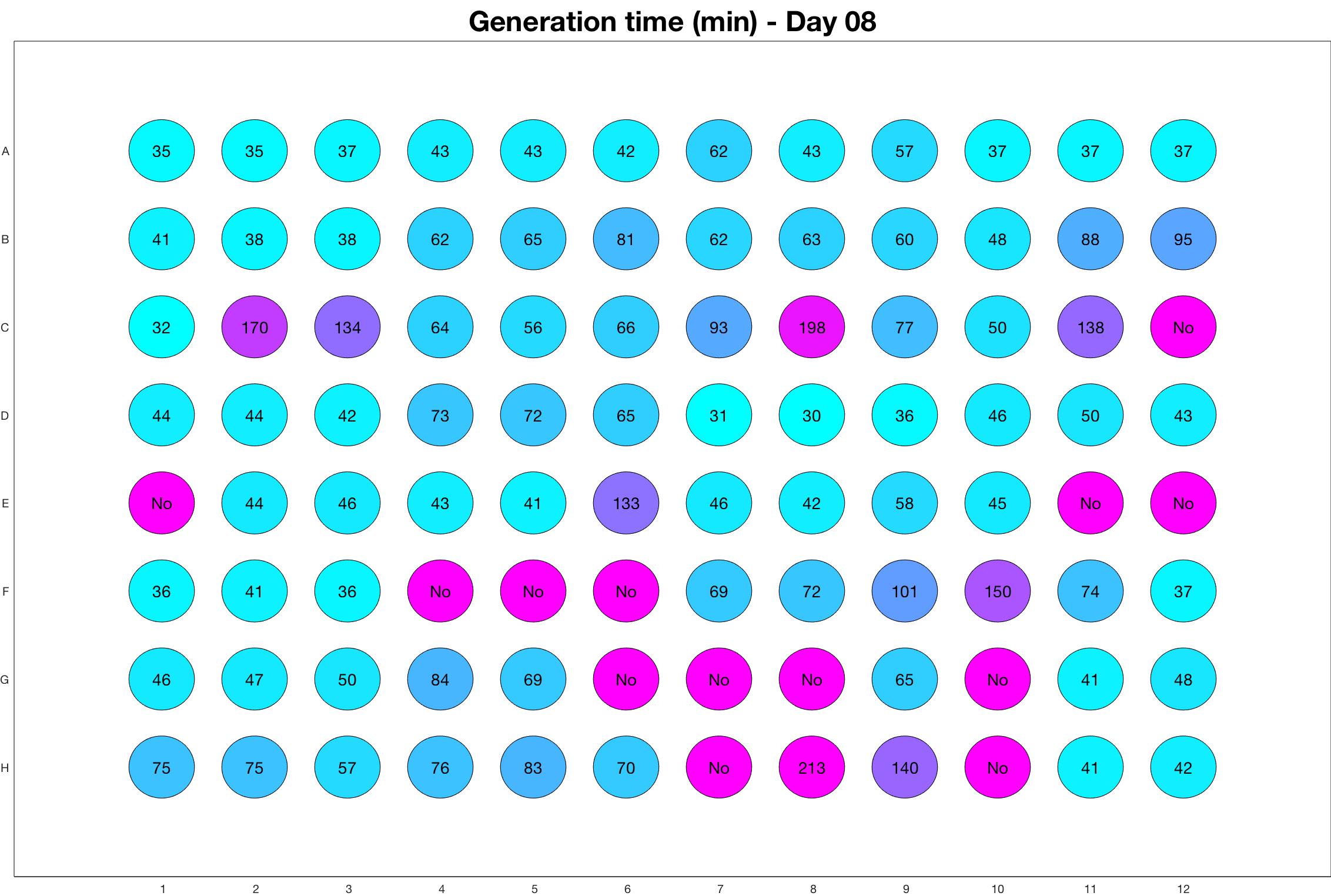
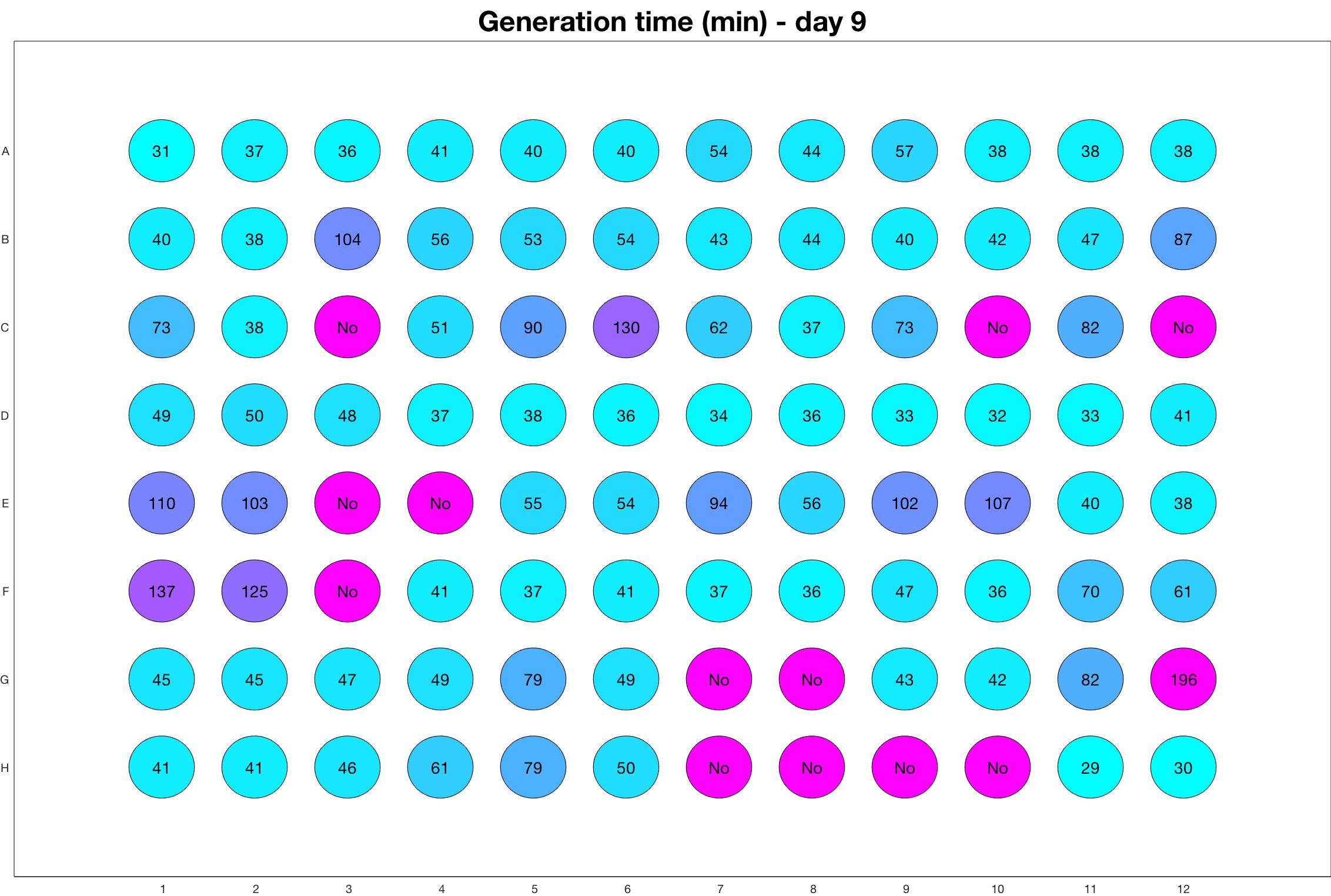
Daily Results
Daily drug regimen
The drug dosing regimen appears below and can be updated by participating student classes (by 3pm, Israel time). The measured generation time calculated by the first 4hr of growth will be updated daily.
Observed doubling time
Sequencing Results
Final project results include the minimal inhibitory concentrations, sequencing of selected genes, and full genome sequence of the winning super-bug.Whole Genome sequencing
We sequenced the entire genome of a single colony (#10) from well C10 to identify the mutations associated with multiple drug resistance. The sequencing uncovered 3 inactivating mutations in ompR (insertion of 4 bases) and alaW (large deletion). In addition we identified a non-synonymous mutation ratA (position 119 in the protein becomes proline instead of leucine).
The wild-type and mutated sequences of ompR and ratA are available HERE.
Sanger sequencing:
Mutations in the gyrase A (gyrA) gene are associated with resistance to Ciprofloxacin. Mutations in the elongation factor fusA are associated with resistance against Kanamycin. Use Blast2Seq tool to align the DNA sequences of the ancestor strain and evolved strains to identify the mutations in these genes.
- Ancestor strain - fusA gene from ancestor strain (fusA_wt.txt)
- Evolved B11 well - fusA sequence of mixed population (B11_Population_fusA_rev.txt)
- Evolved G2 well - fusA sequence of mixed population (G2_Population_fusA_rev.txt)
- Ancestor strain - gyrA gene from ancestor strain (gyrA_wt.txt)
- Evolved H11 well (no mutation found) - gyrA sequence of mixed population (H11_Population_gyrA_fwd.txt)
Note: only 2 mutations were identified in 40 Sanger sequencing reactions we performed. These results indicate that most resistance mutations are likely in different genes than gyrA and fusA. This hypothesis can be directly tested by sequencing the entire genome of evolve strains.


